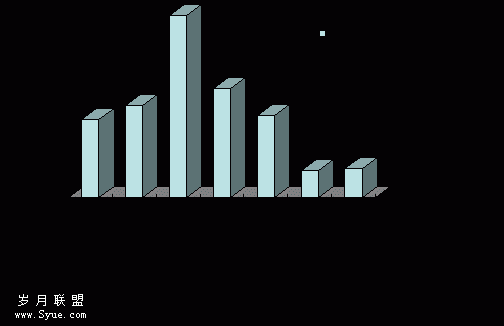Ⅱ类抗原反式激活因子与HLA?DR抗原的关系及其意义
【摘要】 本研究探讨Ⅱ类抗原反式激活因子(CIITA)和人类白细胞抗原(HLA?DR)表达时相的关系和差异,及STAT1?α反义寡核苷酸(STAT1?α AS)对CIITA和HLA?DR的抑制作用。分离健康志愿者外周血T淋巴细胞,给予不同剂量干扰素?γ(IFN?γ)后,用RT?PCR法检测CIITA mRNA,Western blot分析HLA?DR抗原表达,然后给予不同浓度STAT1?α AS和STAT1?α寡核苷酸有义链(STAT1?α S),再次检测CIITA mRNA和HLA?DR的表达。结果表明: CIITA mRNA在IFN ?γ作用后5小时开始表达,14小时达峰值;HLA?DR在28小时后可被检测出,52小时达高峰。5、10和20 μmol/L STAT1?α AS作用于细胞后,CIITA mRNA的表达显著低于对照组(P<0.01),而在S组明显高于AS处理组(P<0.01),S组与对照组间无显著差异;HLA?DR的表达可被STAT1?α AS抑制,AS组仅为对照组的64.3%(P<0.01),S组与对照组间仍无差异; STAT1?α AS作用后,HLA?DR变化同CIITA。结论: CIITA mRNA表达与HLA?DR表达呈正相关且早于后者; STAT1?α AS可特异性抑制CIITA和HLA?DR的表达,并能预防T淋巴细胞激活, CIITA在移植免疫病因中起重要作用。
【关键词】 Ⅱ类抗原反式激活因子 HLA?DR STAT1?α 反义寡核苷酸 干扰素?γ
This work was supported by National Natural Science Fundation of China(国家基金)(No 30170389)and the 135 project fundation of Jiangsu province The class II transactivator (CIITA) was initially cloned using genetic complementation of the in vitro derived HLA?DR?negative mutant B cell line, RJ2.2.5[1]. All current evidence shows that CIITA is absolutely essential for MHC class II gene expression. In addition to regulating the transcription of class II genes, CIITA transfected into nonprofessional antigen?presenting cells (APCs) results in the expression of HLA?DM and invariant chain [2]. CIITA is thus a key regulatory transcription factor to coordinate expression of genes required for HLA class II?mediated immune responses[3]. We investigate the relationship between CIITA and HLA?DR expression in T cells, and explore its potential effect and significance.
Human peripheral blood mononuclear cells (PBMNC) were isolated from heparinized peripheral blood of normal donors using Ficoll?Hypaque density?gradient centrifugation and washed in Hanks' balanced salt solution and used for experiments. To obtain a monocyte?enriched cell population, PBMNC were incubated at 37℃ for 2 hours in plastic Petri dishes in RPMI 1640 culture medium (Gibco) containing 100 U/ml penicillin, 100 U/ml streptomycin and 10% heat?inactivated fetal calf serum (FCS). Nonadherent cells were removed by washing the dishes with prewarmed (37℃) culture medium. Then adherent cells were rinsed off with cold PBS and resuspended in culture medium. CD3?positive cells accounted for 70% of the nonadherent cell population. Interferon?γ (IFN?γ, Shanghai Clone High?Tech Company, China) was added to T cells (6×106/ml) cultures at different final concentration of 50 U/ml to 1200 U/ml for induction of HLA?DR antigen at 37℃, 5% CO2. T cells were harvested respectively at 2, 5, 8, 11, 14, 17, 20, 23 hours to test the expression of CIITA mRNA, and HLA?DR protein were detected respectively at 12, 20, 28, 36, 44, 52, 60, 68, 76 hours after IFN?γ treatment. Samples were stored at -70℃ and were thawed only once.
Reverse transcription?polymerase chain reaction (RT?PCR)
Total RNA was isolated using TRIZOL reagent (Shanghai Bio?Project Company, China) following instructions supplied by the manufacturer. Single?stranded cDNA was synthesized from 2 μg of total RNA in a 40 μl reaction mixture containing 0.2 μg random primer and 300 U reverse transcriptase (Promega, Madison, WI, USA). A 471?bp CIITA fragment and a 306?bp GAPDH fragment were amplified as follows. Five microliters of cDNA was subjected to PCR for 30 cycles, 30 sec of denaturation at 94℃, 1 min of annealing at 61℃ and 1 min of extension at 72℃. Upon completion of the PCR reaction, PCR products were analyzed by ethidium bromide staining in 1.7% agarose gel. Amplification of GAPDH mRNA was used as a control. The sequences of oligonucleotide primers used in PCR amplifications were as follows: CIITA sense, 5'?CAA GTC CCT GAA GGA TGT GGA?3'; CIITA antisense, 5' ?ACG TCC ATC ACC CGG AGG GAC?3'; GAPDH sense, 5'?CGG AGT CAA CGG ATT GGT CGT AT?3'; and GAPDH antisense, 5'?AGC CTT CTC CAT GGT GGT GAA GAC?3' (Synthesized by Invitrogen, USA). In certain cases, serial dilutions of input cDNA were examined to ensure that any effects of CIITA on mRNA induction were not obscured due to a plateau effect in the PCR reaction.
Restriction analysis
PCR products were purified by Wizard DNA purification kit(Promega, USA). PCR?purified products were digested by restriction endonuclease BamHⅠat 37℃ for 4-5 hours. Then the products were analyzed by ethidium bromide staining in 1.7% agarose gel, and self?quantitation was performed by total lab software.
Western blot analysis
Following treatment with IFN?γ, cells were solubilized on ice for 15 minutes in lysis buffer containing 10 mmol/L HEPES(pH 7.9), 10 mmol/L KCl, 0.1 mmol/L EDTA, 0.1 mmol/L EGTA, 1 mmol/L dithiothreitol (DTT), 1 mg/L leupeptin, 1 mg/L apro?nitin, and 10% NP?40. Lysates were vortexed and centrifuged at 1110×g for 10 minutes at 4℃. The supernatants were boiled at 100℃ for 5 minutes in SDS?PAGE sample buffer. The proteins were separated on 7.5 % SDS?PAGE and were electrotransferred onto nitrocellulose membrane (Amersham, Buckinghamshire, UK). The membrane was probed with the mouse monoclonal anti?HLA?DR IgG antibody (Neomarkers, Fremont, CA, USA) overnight at 4℃. For detecting bound antibodies, alkaline phosphatase?conjugated goat anti?mouse IgG (Sigma) and nitro?blue tetrazolium (NBT)/5?bromo?4?chloro?3?indolyl?phosphate (BCIP) color substrate (Promega) were used. Then the bands on the membrane were scanned and analyzed with an image analyzer (LabWorks software, UVP upland, CA, USA).
STAT1?α antisense oligonucleotides treatment of T cells
The antisense STAT1?α oligonucleotide (STAT1?α AS), CCACTGAGACATCCTGCCAC, was complementary to the region of STAT1?α mRNA containing the AUG codon. At the same time, the sense STAT1?α oligonucleotide (STAT1?α S), GTGGCAGGATGTCT?CAGTGG, was used as control. T cells were mixed with oligonucletide (5 μmol/L) and incubated for 6 hours at 37℃. Medium was removed and replaced with RPMI 1640 containing 10% FCS and 1000 U/ml IFN?γ. Then the experiment was divided into three groups: AS group, S group and control group. In control group medium only contained 1000 U/ml IFN?γ and RPMI 1640, not incubated with oligonucletide. T cells were harvested and analyzed for CIITA mRNA by RT?PCR at 14 hours, or cells were lysed and analyzed for HLA?DR protein at 52 hours as described above.
Statistics
All experiments were repeated four times with different batches of cells. Values were expressed as mean±SD of four independent experiments. Statistical analysis of the results was performed by one?way analysis of variance followed by the Duncan's new multiple range method. P<0.05 was considered as significant.
Results
Restriction analysis
The length of PCR products of CIITA gene is 471 bp. The products contained one BamHⅠ site. The PCR products of CIITA gene were digested by BamHⅠ, and two segments were obtained after digestion, one was 163 bp and the other was 308 bp. The bands of the PCR products and the fragments after digestion were shown in Figure 1.
Figure 1. CIITA gene PCR products before digestion and after digestion. M: marker. Lane 1: bands after enzymolysed by BamH1 (308 bp and 163 bp). Lane 2: PCR products (471 bp)
Expression of CIITA mRNA in T cells
Expression of CIITA was positively correlated with the concentrations of IFN?γ when the levels of IFN?γ were between 100 U/ml and 1000 U/ml. When the concentration of IFN?γ was 1 000 U/ml, the fluorescence of CIITA /GAPDH was the strongest and the expression of CIITA was no longer increased when the levels of IFN?γ were above 1 000 U/ml. Therefore 1 000 U/ml of IFN?γ was chosen as the optimum dose (data not shown). We initially analyzed T cells for expression of CIITA mRNA. T cells were constitutively negative for CIITA mRNA, but the expression was induced by IFN?γ in a time?dependent manner (Figure 2). CIITA mRNA was detectable at 2 hours after IFN?γ treatment and reached the peak at 11 hours, then declined gradually, but the expression of CIITA was still evident at 20 hours after IFN?γ stimulation.
Figure 2. Expression of CIITA and GAPDH at various time points after 1 000 U/ml IFN?γ treatment. Lane 1-8: 2, 5, 8, 11, 14, 17, 20, 23 hour groups; M: marker. CIITA gene products were 471 bp, GAPDH was 306 bp.
Expression of HLA?DR protein in T cells
T cells were constitutively negative for HLA?DR, but could be induced to express by IFN?γ in a time?dependent manner. IFN?γ induced HLA?DR protein was detectable after a 28?hours stimulation with IFN?γ, the strongest expression was found after a 52?hours exposure to IFN?γ, then declined gradually, however HLA?DR antigen was still detectable at 76 hours after stimulation (Figure 3). CIITA expression was positively correlated with HLA?DR expression, and was detectable earlier than that of latter.
Figure 3. Expression of HLA?DR after 1000 U/ml IFN?γ treatment at various time points (hour).
Inhibition of CIITA and HLA?DR expression by antisense oligonucleotides against STAT1?α
There was no significant difference in the expression of CIITA mRNA between the AS group and control group when the concentration of AS was 1 μmol/L. The expression of CIITA mRNA was significantly lower in the AS group than that in the control group at 5, 10 and 20 μmol/L (P<0.01), but there was no significant difference in CIITA expression between 10 μmol/L AS group and 20 μmol/L AS group (P>0.05). The expression level of CIITA mRNA in the AS group was about 63.3% of that in the control group (P<0.01). The expression of CIITA mRNA in the S group was higher than that in the AS group (P<0.01), but there was no siginificant difference between the S group and the control group (P>0.05)(Table).
Table . Expression of CIITA mRNA in AS group, S group and the control group※at 14 hours (±SD)
GroupNumber of samplesOD ratio(CⅡTA/GAPDH)AS40.475±0.039*▲S40.749±0.037#Control40.750±0.062※AS group , S group at 5 nmol/L, *P<0.01, compared with control group, ▲P<0.01, compared with S group, #P>0.05, comapred with control group.
The expression of HLA?DR antigen was significantly inhibited by STAT1?α AS, and the expression level of HLA?DR protein in the AS group was about 64.3% of that in the control group(P<0.01), while there was no significant difference in HLA?DR expression between the S group and the control group (P>0.05) (Figure 4A). According to OD, statistical analysis of Western blot was displayed in Figure 4B, which also showed that STAT1?α AS could markedly inhibit the expression of HLA?DR antigen. In addition, the changes in HLA?DR expression were very similar to those in CIITA expression after STAT1?α AS treatment.
Discussion
Acute graft verse host disease (aGVHD) occurs when transplanted donor?derived T cells recognize and react to histoincompatible recipient antigens and cells. Final consequences of aGVHD process are widely variable. GVHD is a major obstacle to allogeneic hematopoietic stem cell transplantation (HSCT), leading to a significant morbidity and mortality. aGVHD mostly is a consequence of damage to host tissues by activating donor?derived T lymphocytes in response to the MHC incom?
Figure 4. Western blot results of HLA?DR expression after STAT1?α antisense oligonucleotide treatment (A) and statistical analysis of the Western blot results (B). ▲P<0.01, compared with control. *P<0.01, compared with S group. #P>0.01, compared with control group.
patibility. A three?phase model elucidates the three major processes that lead to GVHD[4]. The first phase is involved in tissue damage secondary to the conditioning regimen. Activated cells from damaged recipient tissues secrete many inflammatory cytokines, such as interleukin?1 (IL?1), tumor necrosis factor?alpha (TNF?α), granulocyte?macrophage colony?stimulating factor (GM?CSF), and interferon?γ (IFN?γ). Dysre?gulated release of the cytokine may upregulate the adhesion molecules and enhance the recipient MHC antigens. Increased expression of donor?tissue antigens may augment recognition of histoincompatible host?tissue antigens by alloreactive donor T cells. The second phase consists of donor T cell activation, stimulation, and proliferation. Donor T cell activation requires two signaling events[5]. TCR?peptide?MHC interaction, and a lattice formation between allopeptide bound to host MHC and donor T cell receptor is the first signal. This process is limited by MHC. The second, i.e., costimulatory signal , is provided by APCs and requires cell?to?cell contact. Finally, the effect phase comprises the third phase of GVHD pathophysiology. The first phase of GVHD can not be avoided, however we might interfere the activation of T cells or can detect some markers before T cell activation, so that we can disturb the occurrence of GVHD or discover GVHD early.
HLA?DR antigen is a kind of MHC class II antigens, and plays a key role in GVHD. HLA?DR also is a marker of activated T cells. CIITA is a master regulator essential for both constitutive and IFN?γ?inducible MHC class II expression, and initiated a "limitation factor" to regulate the expression of MHC class II antigens[6]. So it is necessary to study the correlation and difference of expression phase between CIITA and HLA?DR antigens, which will help to investigate the potential effect and significance of CIITA in GVHD.
It is noteworthy that some reports have described none of HLA?DR antigens on the surface of normal T cells, but the increased expression of these molecules can be found under some conditions, especially after IFN?γ treatment. Since a long lag period is required after IFN?γ stimulation before HLA?DR antigens can be detected (28 hours), and the response is dependent on ongoing protein synthesis, therefore CIITA expression is earlier than that of HLA?DR expression. Together, these data indicated that in the process of IFN?γ?induced HLA?DR antigen expression, IFN?γ might first induce the expression of CIITA, then followed by inducing HLA?DR expression.
To confirm our hypothesis, we selected antisense oligonucleotide to inhibit the expression of CIITA, then we observed the change of HLA?DR expression. CIITA gene contains four promoters, and promoter Ⅳ is necessary for IFN?γ?induced HLA?DR antigen expression. IFN?γ binds to its cognate receptor, resulting in receptor phosphorylation and the docking of the STAT1 protein. STAT1 is in turn phosphorylated, and this modification leads to the formation of a homodimer. The homodimer is then translocated into the nucleus to bind promoter Ⅳ of CIITA that activates gene expression[7]. So, we designed a sequence of 20 nucleotides around the AUG codon of STAT1?α mRNA with a maximal melting temperature and minimal self?complementarity. Phosphorothioate oligonucleotides that exhibit enhanced nuclease resistance were synthesized for this study[8], at the same time the sense oligonucleotides were used to be compared more accurately.
These results clearly demonstrated that CIITA expression was positively correlated with HLA?DR expression, and CIITA might regulate the expression of HLA?DR antigens. Because HLA?DR plays a key role in GVHD so we think that CIITA may also play an important role in GVHD pathophysiology. Some studies have demonstrated that CIITA can inhibit interleukin?4 secretion and promote T cells to differentiate to T helper 1 cells[9,10]. Therefore, the up?regulated CIITA may induce "cytokine storm", then GVHD occurs and develops.
CIITA mRNA can be detectable earlier than HLA?DR. The up?regulated HLA?DR is a early marker of T cell activation, so the up?regulated CIITA may be an earlier marker of T cell activation[11]. CIITA can si?multaneously up?regulate T helper 1 cells then result in GVHD. Thus we think CIITA may be an earlier predicting marker of GVHD.
Because of the accurate regulation of CIITA for MHC class II genes and antigen submitting correlated genes, CIITA plays a key role in transplantation immunity. Many studies on the effects of CIITA in transplantation rejection have been reported, which shows that the reduction of CIITA expression can notably delay transplantation rejection[12]. However, there was no report about the effects of CIITA in GVHD. Our principal studies indicate that CIITA might play an important role in the development of GVHD and may be an earlier predicting marker of GVHD.
【】
1Steiml V,Otten LA,Zufferey M,et al. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency(or bare syndrome lymphocyte syndrome).Cell, 1993;75:135-146
2Ghosh N, Piskuich JF,Wright G,et al. A novel element and a TEF?2?like element activate the major histocompatibility complex class II transactivator in B?lymphocyte. J Bio Chem, 1999; 274:32342-32350
3Chang CH, Gourley TS, Sisk TJ. Function and regulation of class II transactivator in the immune system. Immunol Res, 2002;25:131-142
4Ferrara JL, Levy R, Chao NJ. Pathophysiologic mechanisms of acute graft?verse?host disease. Biol Blood Marrow Transplant, 1999; 5:347-356
5Goker H, Haznedaroglu IC, Chao NJ. Acute graft vs host disease: pathobiology and management. Exp Hematol, 2001; 29:259-277
6Steimle V,Siegrist CA,Mottet A, et al. Regulation of MHC class II expression by interferon?γ mediated by the transactivator gene CIITA. Science, 1994;265:106-109
7Mottet AM,Berardino DW,Otten LA,et al. Activation of the MHC class II transactivator CIITA by interferon?gamma requires cooperative interaction between Stat1 and USF?1. Immunity, 1998;8:157-166
8Lee YJ, Benveniste EN. State1α expression is involved in IFN?γ induction of the class II transactivator and class II MHC genes. J Immunol, 1996;157:1559-1568
9Otten LA, Tacchini?Cottier F, Lohoff M, et al. Deregulated MHC class II transactivator expression leads to a strong Th2 bias in CD4+ T lymphocytes. J Immunol, 2003;170:1150-1157
10Sisk TJ, Gourley T, Roys S, et al. MHC class II transactivator inhibits IL?4 gene transcription by competing with NF?AT to bind the coactivator CREB binding protein (CBP)/p300. J Immunol, 2000;165:2511-2517
11Holling TM, van der Stoep N, Quinten E,et al. Activated human T cells accomplish MHC class II expression through T cell?specific occupation of class II transactivator promoter III. J Immunol, 2002;168:763-770
12Brickey WJ,Felix NJ,Griffiths R,et al.Prolonged survival of class II transactivator?deficient cardiac allografts. Transplantation, 2002; 74:1341-1348 []?μβαγ[AKU¨] [AKX?]±SD